Marking sections in a script
When R scripts become longer, you’ll find you want to make subdivisions in the scripts so you can easily navigate it, avoiding endless scrolling. You can mark sections by creating a section by starting a comment like normal (#), typing a section tile and following that by a series of ‘-‘ signs like this:
# Section title -------
This allows you to fold your code (hide code, showing only the section header), or jump through the script Code folding:
Section navigation:
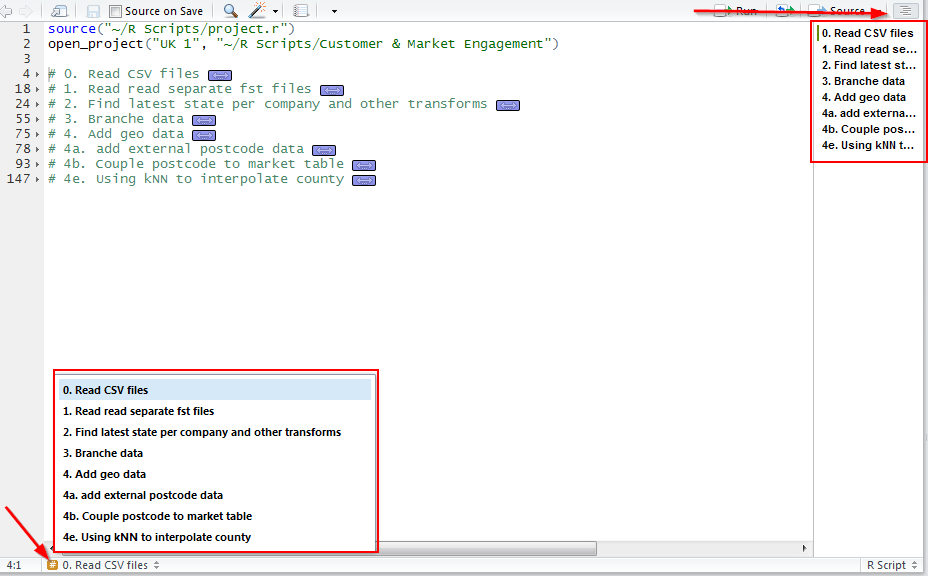
Calling scripts from scripts
Whenever you tend to re-use script in several instances, you might want to place it in a seperate file you can call from within the script you intend to execute. You can call a script using the source(filename) function.
This could be employed, for example, when you create multiple analysis scripts on the same set of input files. Frequently, reading and transforming data is generic for all analyses. Data needs to be read and transformed to it meets R needs: dates need to be R dates, numbers need to be recognized as such, values might need to be factored, empty values need to be handled and so on. It makes sense to put all this data-set related statements in a separate script. Calling the script in the analysis script will result in the creation of variables, data-sets and functions so they are made available in the analysis script itself.
An example of script in script
I prefer to keep the bulk of my data processing out of my R Markdown document, so my analysis isn’t cluttered by code. For this reason I make R script files for each source file, so I can easily locate where I should adjust what in case something is not how I wanted things to be. All the files for each data source is called from a central data-prep file, wherein I do all data transformations that are cross-source. This idea is depicted in the diagram below:
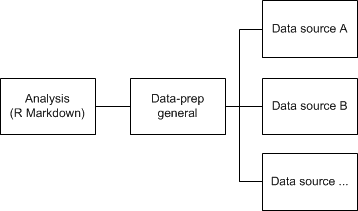
In each data-prep file I don’t do the data processing directly, instead I create a function containing all data processing. Why do I do this? I might not want to do all data-processing again and again every day I restart my computer, but instead load all pre-processed data. The skeleton of that function looks something like this:
prep_datasource_A <- function(do_processing){
if(process_data){
# Load raw data
# Transform raw data
# Write processed file
} else {
# Load previously processed data
}
}
As you can see the function takes a boolean process_data. The value of this parameter determines whether we process the data from scratch or whether we load pre-processed data.
Setting working directories
When working on a project, you probably want all your files to be in a folder you made for that project. If you set the working directory in a script, all following file calls are presumed to be files within that directory.
- setwd(dir) - setting the working directory
- getwd() - getting the current working directory
The default directory is your ‘My Documents’ directory. When changing the working directory, keep in mind that R uses foward slashes (/) instead of the Windows standard of backward slashes (). If the working directory is a subfolder of your ‘My documents’ folder use the ~ as the start of your working directory call to setwd; in this way other users can use the exact replica of the script as long as they use the same folder conventions. And example:
setwd("~/R scripts/My Project/")