When I do larger projects I always encounter three problems:
- My analysis script becomes cluttered with heaps and heaps of code. I prefer to keep the bulk of my data processing out of my R Markdown document, so my analysis isn’t cluttered by code.
- Whenever a script gets larger and larger, it also becomes more and more difficult to track down bugs.
- Also I’d hate to do all data-processing again and again every day I restart my computer when I continue working on my project. It takes up a lot of time. I don’t mind getting coffee while my computer works, but it becomes problematic when my computer is still crunching after I’ve finished my coffee.
To address these problems this I made a framework which I’ll explain in this tutorial.
Project scripts structure
To ensure I don’t get one blob of code, I make R script files that will do data loading and transforming for each source. This way I can easily locate where I should adjust code in case something is not how I wanted things to be. All the files for each data source is called from a central data-prep file, wherein I do all data transformations take place that are cross-source. This idea is depicted in the diagram below:
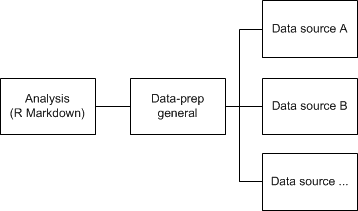
As a side effect this approach let’s you recycle the data prep scripts for source files for different projects using the same source data. In the following sections I’ll discuss the scripts for data-source prepping, general prepping and the final analysis script. I’ve also created a working example with nonsensical data you can download here.
Data source prep scripts
In each data-prep script for a source, data processing isn’t processed directly, instead all data processing is put inside a function. This function takes a boolean process_data. The value of this parameter determines whether we process the data from scratch or we load pre-processed data. The skeleton of that function looks something like this:
prep_datasource_a <- function(do_processing){
if(do_processing){
# Load source data
# Transform source data in data frame 'df_source_a'
# Write dataframe 'df_source_a' to processed file
write.fst(df_source_a, "source_a.fst", 100)
} else {
# Load previously processed data in a data frame 'df_source_a'
df_source_a <- read.fst("source_a.fst")
}
stopifnot(exists("df_source_a")) # If 'df_source_a' doesn't exists, stop the script
return(df_source_a)
}
I use the fst library for processed file storage and retrieval since it is fast and creates compact files. I explain fst’s usage here.
General data-prep script
In the general data-prep script, which I’ll call data-prep.R, the data source scripts are loaded, which looks something along the lines of this:
source("prep_datasource_a.R")
source("prep_datasource_b.R")
source("prep_datasource_c.R")
Then another function is created that looks quite similar to the prep_datasource_a function, but it first calls all data source prep functions. Depending on the value of the do_processing parameter the general data-prepping is done, or a pre-prepped file is loaded. Then a list is created with all the data frames, so all can be returned to the analysis script.
prep_datasources <- function(do_processing){
prep_datasource_a(do_processing)
prep_datasource_b(do_processing)
prep_datasource_c(do_processing)
if(do_processing){
# Create cross data source in data frame 'df_source_general'
# Write dataframe 'df_source_general' to processed file
write.fst(df_source_general, "source_general.fst", 100)
} else {
# Load previously processed data in a data frame 'df_source_general'
df_source_general <- read.fst("source_general.fst")
}
stopifnot(exists("df_source_general")) # If 'df_source_general' doesn't exists, stop the script
# Create a list with all data frames and return it
list_df <- list(df_source_a = df_source_a,
df_source_b = df_source_b,
df_source_c = df_source_c,
df_source_general = df_source_general)
return(list_df)
}
Analysis script
In the analysis script, be sure to point the working directory where it contains all scripts. The general data prep script is loaded, which in turn will load all data source prepping scripts. The do_processing variable controls whether the data-prepping is done from scratch or the pre-prepped data is loaded from files. The prep_datasources function is called with the do_processing variable as a parameter from within the list2env function. The list2env function will unlist the data frame list and put all the data frames in the environment.
setwd("/home/mark/R Scripts/structuring-data-processing/")
source("data-prep.R")
do_processing <- FALSE
list2env(prep_datasources(do_processing), .GlobalEnv)
Now you’re good to go doing your analysis!
A working example is downloadable here.
• 0 Comments